I pick mushrooms, grow them, and occasionally eat them, while using them to solve current problems and future challenges
François Gaascht, PhD

Hello,
I am François. I can usually be found with one foot in the field, collecting mushrooms to add more specimens to my own field-to-laboratory Basidiomycota strain library—such as this 3 kg giant puffball (Calvatia gigantea)—one foot in the laboratory, running exciting experiments to investigate and valorize Fungi from my biobank by discovering bioactive natural products and biocatalysts.
I am currently a Research Associate and Laboratory Manager in Claudia Schmidt-Dannert’s Laboratory at the BioTechnology Institute (University of Minnesota, Minnesota, USA). My research focuses on building a field-collected Basidiomycota strain library and screening it for the discovery and expression of gene clusters and biocatalysts for industrial applications. I use bioinformatics approaches to mine the genomes of unsequenced fungi I collect, discovering new gene clusters and economically important genes that I clone for heterologous expression in different microbial chassis (Bacteria or Yeasts).
I am particularly interested in how mushrooms can be used to solve current problems and future challenges, such as the discovery of new therapeutics (antibiotics, anticancer, antimalarial, antiviral agents), bioremediation, biofuel production, textile innovation, biomaterials, and even space exploration.
I am always happy to connect with people and share my ideas and expertise. Feel free to contact me via email or on social media.
Education
- PhD Doctor Europaeus in Health and Life Sciences (University of Lorraine, France)
- M.Sc. in Molecular and Structural Biology (University of Strasbourg, France)
- B.Sc. in Biochemistry and Molecular Biology (University of Strasbourg, France)
Experience
- Research Associate - Laboratory Manager (University of Minnesota, USA), 2022 - Current
- Postdoctoral Research Associate (University of Minnesota, USA), 2016 - 2021
- Chemist in Natural Product Chemistry (Fundación MEDINA, Spain), 2014 - 2015
- PhD Student - Early Stage Researcher (ESR) in MSCA (Marie Skłodowska-Curie Actions) RedCat Innovative Training Network (ITN) (Laboratoire de Biologie Moléculaire et Cellulaire du Cancer, Luxembourg), 2009 - 2013
My expertise
My Research Interests
- Discovery and identification of bioactive natural products for therapeutic applications.
- Genome mining of unique Basidiomycota strains and identification of new biocatalysts.
- Expression of fungal biocatalysts for industrial and biotechnological applications.
- Engineering and optimization of non-model fungal strains.
From the Fungi to the biomolecules...
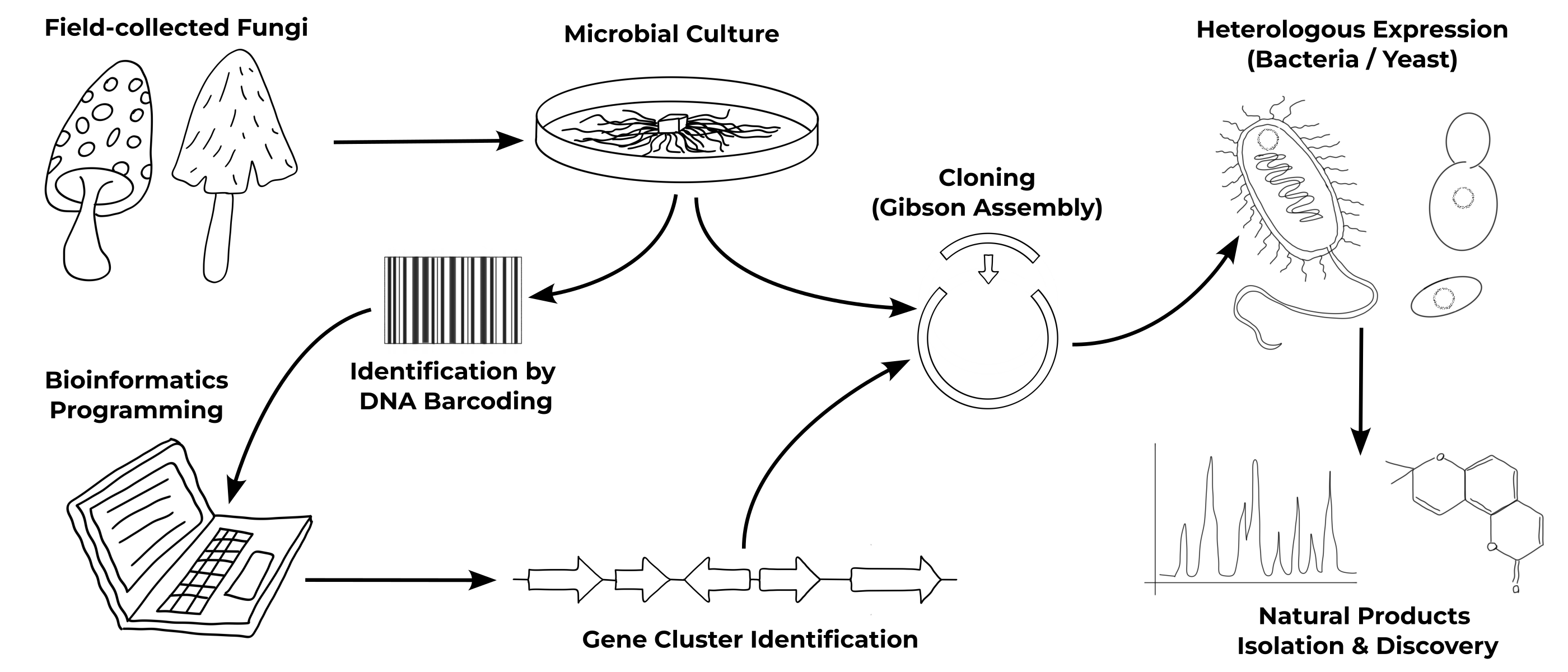
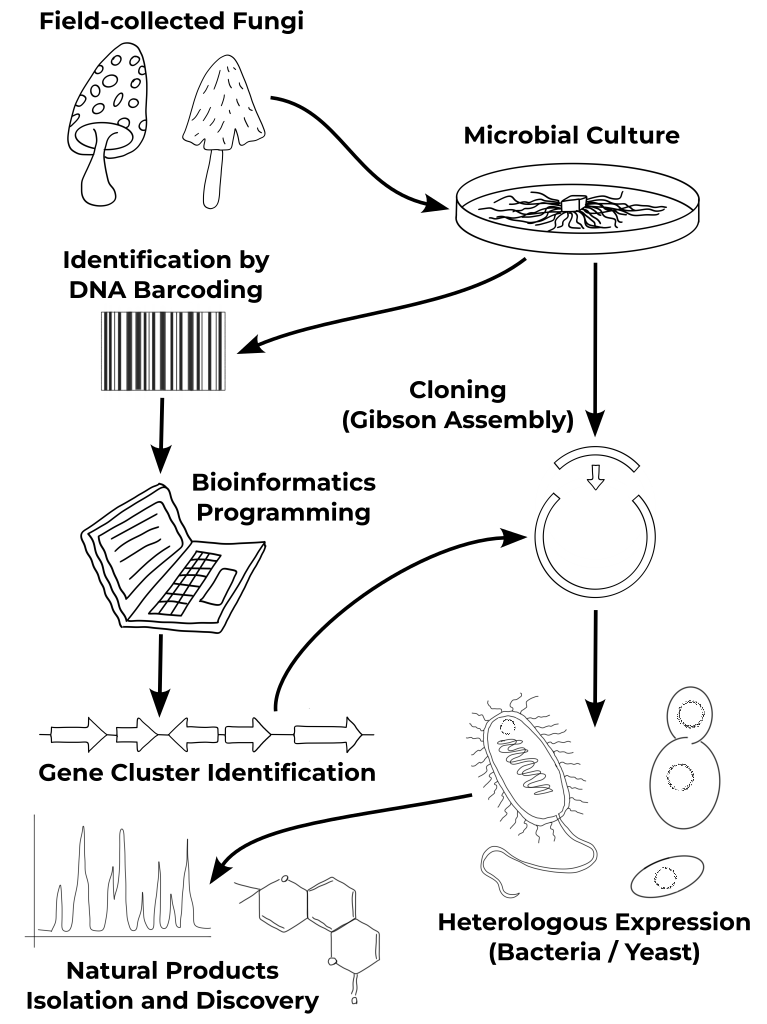
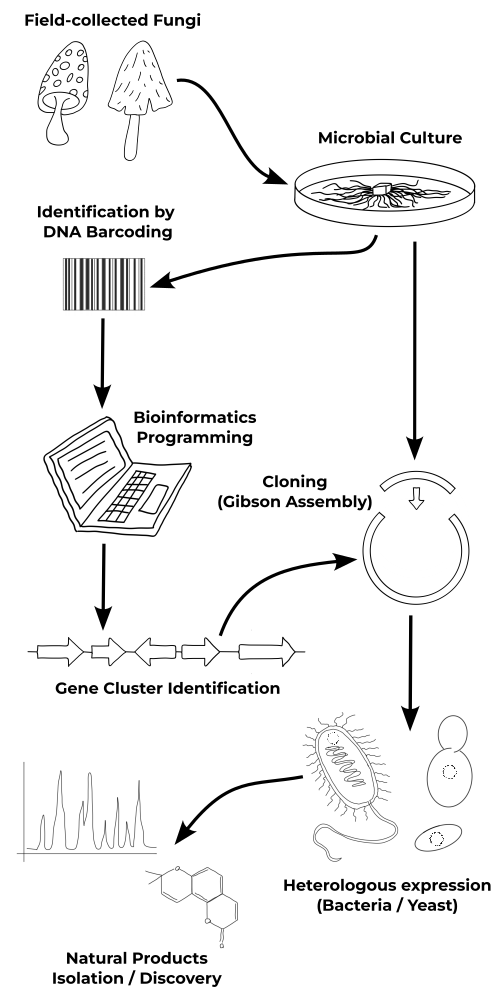
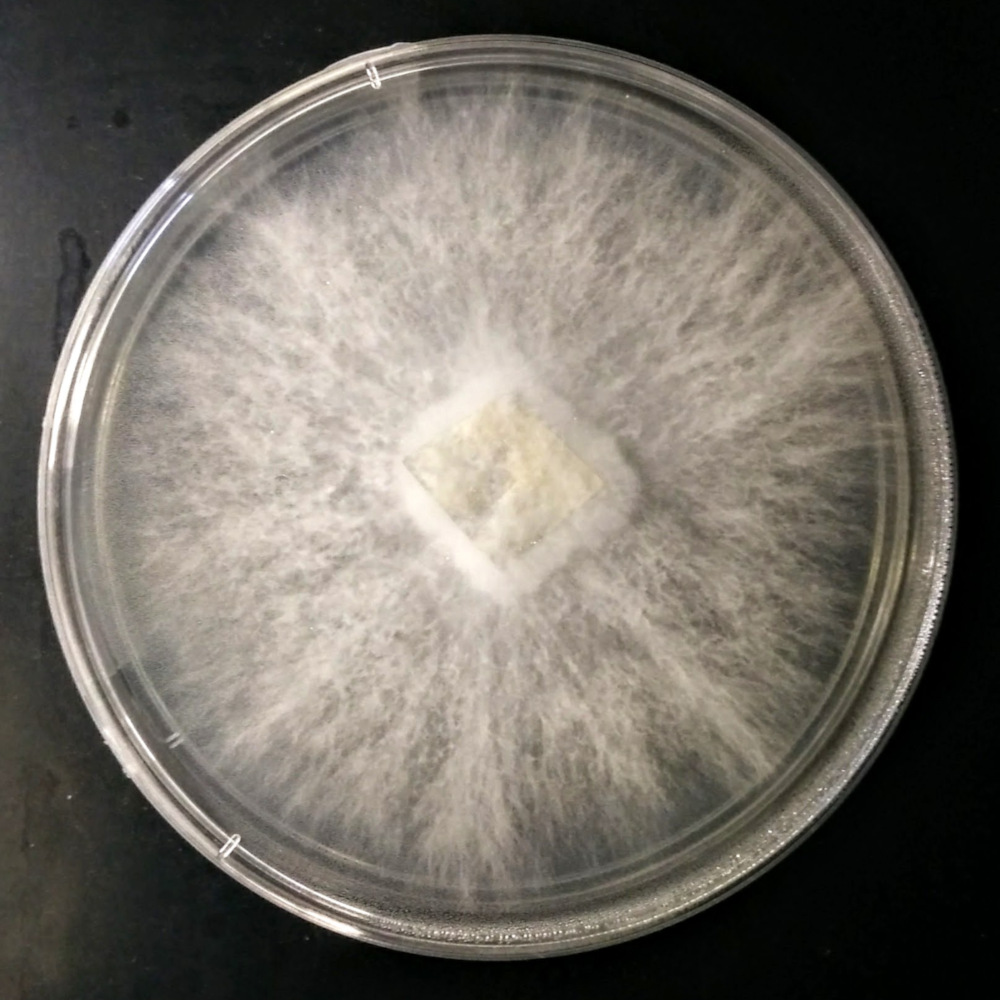
Fungi Collection & Identification
Over the past eight years, I have developed expertise in the collection of fungi, mainly Basidiomycota, their establishment into microbial cultures, and identification by DNA barcoding using appropriate primers. By exploring different biotopes and environments in Minnesota (USA), I created a unique Basidiomycota strain library with more than 50 specimens, including several species with different strains from various locations.
- Collection of living specimens (Basidiomycota and Streptomyces).
- Establishment of microbial cultures.
- Identification of species and strains by DNA barcoding from fruiting bodies, liquid, or solid cultures.
- Creation of a field-to-laboratory Basidiomycota strain biobank (over 50 strains).

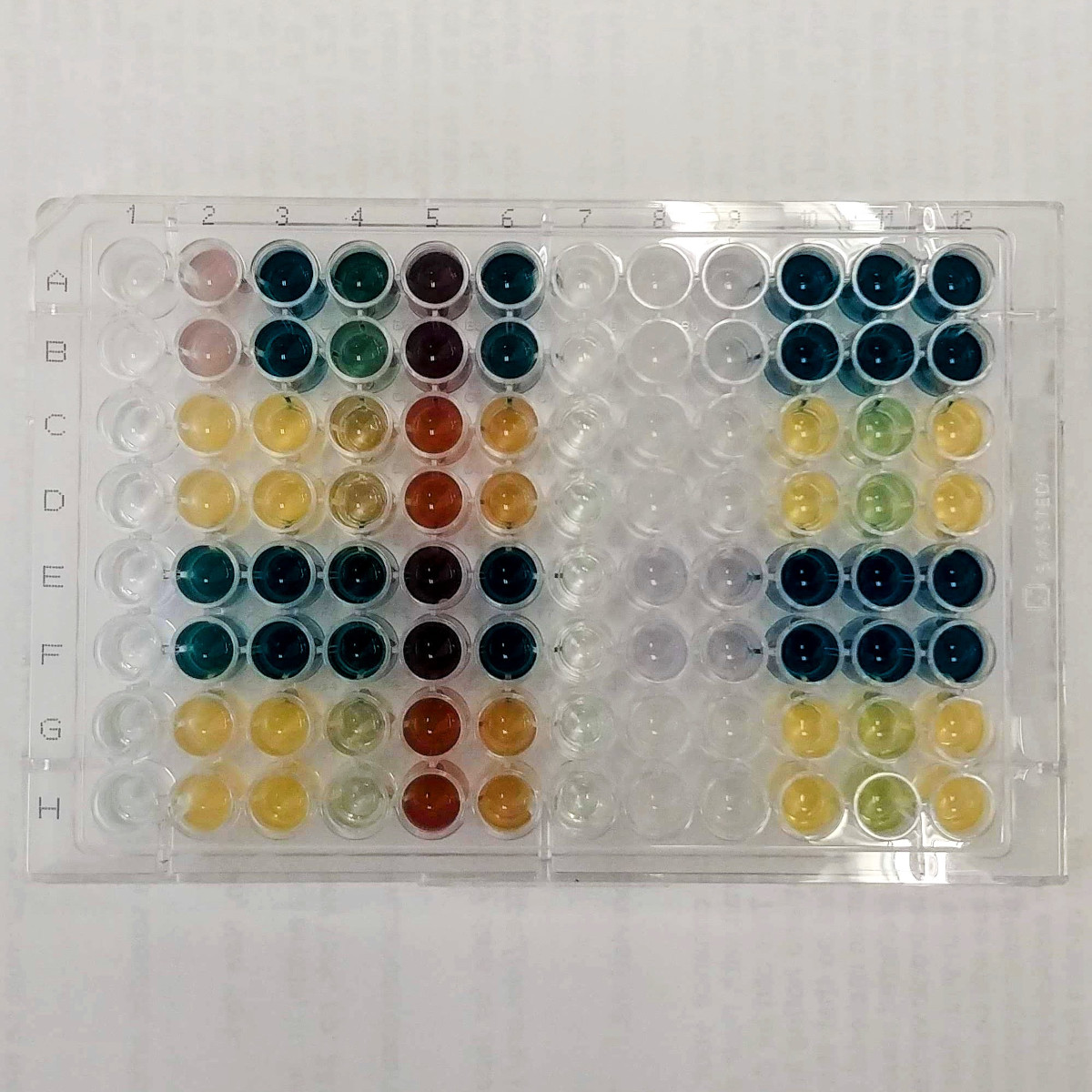
Natural Products Chemistry
My work and interest in natural product discovery have driven me to develop strong skills in the isolation and identification of bioactive secondary metabolites from microbial cultures.
- Fermentation in formats ranging from 96-deep well plates (1.2 mL) to large flasks (2 L).
- Preparation of extracts from microbial fermentations.
- Development of analytical methods using HPLC.
- Detection, quantification, and monitoring of secondary metabolites (HPLC, LC-MS).
- Isolation of compounds through liquid-liquid extraction, solid-phase extraction, flash chromatography, and (semi-) preparative HPLC.
- Structure elucidation using high-resolution mass spectrometry (HR-MS) and 1D/2D NMR spectroscopy (1H, 13C, COSY, HSQC, HMBC).
- Discovery and identification of volatile natural products (terpenes) from fungal fermentations using GC-MS.
Screening | Cell Biology
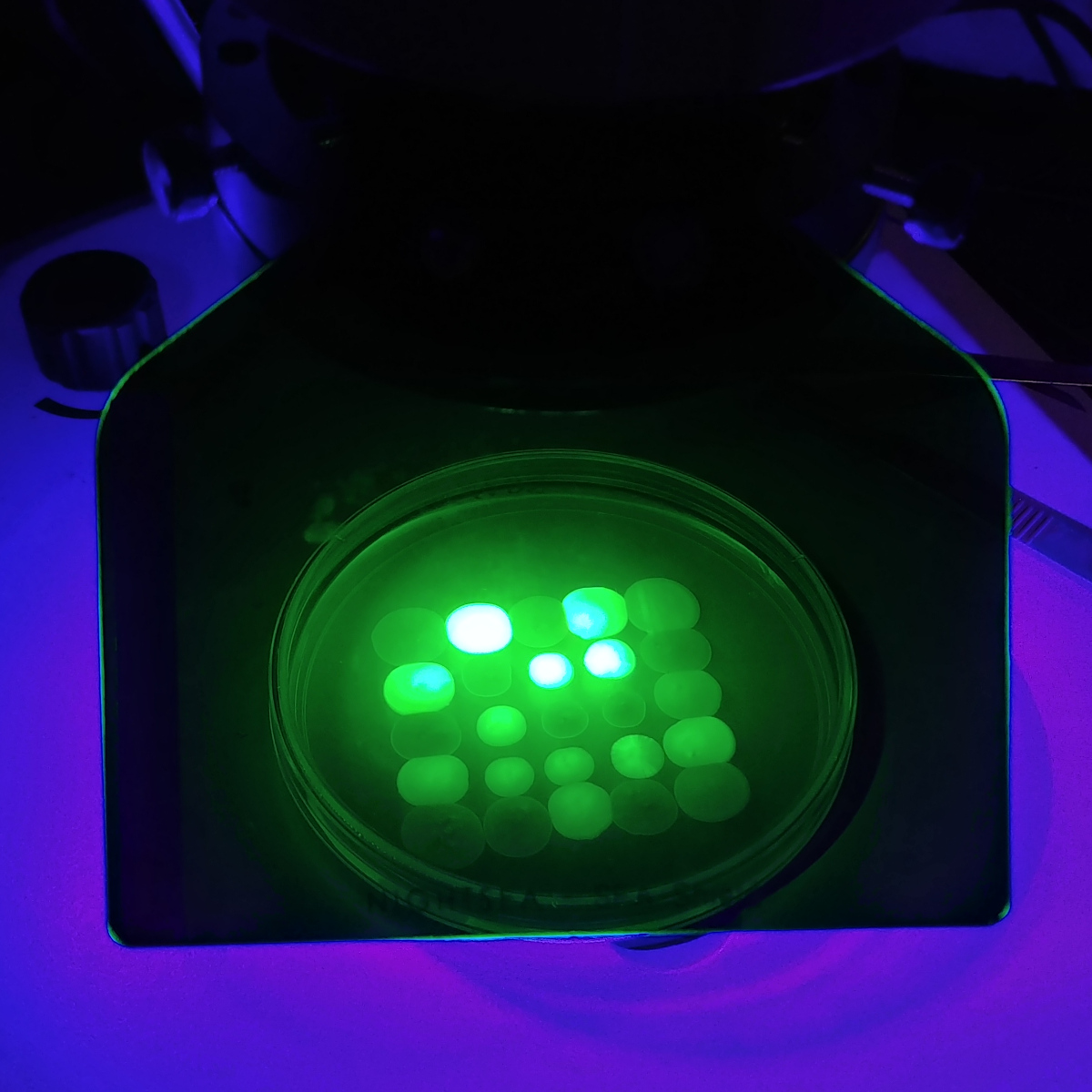
I began my scientific career as a cancer biologist and have since gained extensive experience in the discovery and characterization of natural products from various origins (plants, semi-synthetic sources, and marine fungi) with applications as anticancer or antimicrobial agents.
- Human cancer cell culture (adherent and suspension cell lines).
- Development of protocols and bioassays for large-scale screening.
- Optimization and miniaturization of protocols (96-well plates).
- Cytotoxicity assays (manual methods and commercial kits) for anticancer activity discovery and identification of cell death mechanisms.
- Screening of fungal fermentations for enzymatic activity (e.g., lignocellulolytic, laccase, and peroxidase activities).
- Quantitative real-time PCR (qRT-PCR).
- Flow cytometry.
- Fluorescence microscopy.
- SDS-PAGE and Western blotting.
Synthetic Biology
After mining the genomes of several specimens from my Basidiomycota fungal strain library to isolate genes of interest, I have developed strong expertise in the isolation, cloning, and expression of genes with industrial and economic significance in appropriate heterologous hosts.
- DNA/RNA isolation from fungal cultures.
- Polymerase chain reaction (Touchdown PCR, Gradient PCR, Colony PCR, Multiplex PCR, 2-step PCR).
- Isolation of genes of interest from unsequenced and unique fungi.
- Sequence design, DNA assembly, and plasmid creation.
- Molecular cloning (restriction enzyme-free) of large and/or multiple fragments using Gibson assembly.
- Development, optimization, and simplification of complex protocols and SOPs.
- Yeast homologous recombination.
- Constitutive or inducible expression of intracellular or secreted proteins and biocatalysts.
- Expression in appropriate model and non-model organisms, including Escherichia coli, Pseudomonas putida, Deinococcus radiodurans, Bacillus subtilis, and the yeast Pichia pastoris.
- Strain engineering and metabolic pathway engineering.

Bioinformatics
The various research projects in molecular biology and biochemistry I have worked on have led me to develop strong bioinformatics and computational biology skills, covering different aspects, such as:
- BLAST and sequence alignment.
- Phylogenetic analysis.
- Genome mining, identification, and description of biosynthetic gene clusters.
- Gene prediction for Basidiomycota phylum.
- Python 3 programming and the BioPython library.
- Protein structure modeling and prediction (PyMOL, SWISS-MODEL, AlphaFold).
- Experience with various software (Aliview, CLC Genomics Workbench, Jalview, MEGA, SnapGene, UGENE).
- Experience with Linux (Ubuntu).
Coding | Programming
I recently decided to learn and develop coding and programming skills in R, Python 3, HTML5, and CSS3. Using online resources, my objective was to develop new skills that would help automate recurring tasks, solve complex professional challenges, manage and visualize large datasets, and generate high-quality figures.
R Programming Language (R Studio)
- Data management and analysis of large datasets.
- Data visualization (plots, gene clusters, etc.) and design of high-quality figures for publications and presentations.
Python 3 Programming Language
- Creation and development of smart Python 3 scripts and programs to solve challenging and complex problems, automate time-consuming workflows, and manage and analyze large datasets for biotechnology research projects (e.g., extraction of large datasets from files or online resources and websites).
Python 3 scripts and programs I have completed and am currently working on have been deposited on my personal GitHub profile page.
HTML5 - CSS3 - Bootstrap 5
- Design of this responsive website from scratch using HTML5 and CSS3.
Fungi Collection & Identification
Over the past eight years, I have developed expertise in the collection of fungi, mainly Basidiomycota, their establishment into microbial cultures, and identification by DNA barcoding using appropriate primers. By exploring different biotopes and environments in Minnesota (USA), I created a unique Basidiomycota strain library with more than 50 specimens, including several species with different strains from various locations.
- Collection of living specimens (Basidiomycota and Streptomyces).
- Establishment of microbial cultures.
- Identification of species and strains by DNA barcoding from fruiting bodies, liquid, or solid cultures.
- Creation of a field-to-laboratory Basidiomycota strain biobank (over 50 strains).
Natural Products Chemistry
My work and interest in natural product discovery have driven me to develop strong skills in the isolation and identification of bioactive secondary metabolites from microbial cultures.
- Fermentation in formats ranging from 96-deep well plates (1.2 mL) to large flasks (2 L).
- Preparation of extracts from microbial fermentations.
- Development of analytical methods using HPLC.
- Detection, quantification, and monitoring of secondary metabolites (HPLC, LC-MS).
- Isolation of compounds through liquid-liquid extraction, solid-phase extraction, flash chromatography, and (semi-) preparative HPLC.
- Structure elucidation using high-resolution mass spectrometry (HR-MS) and 1D/2D NMR spectroscopy (1H, 13C, COSY, HSQC, HMBC).
- Discovery and identification of volatile natural products (terpenes) from fungal fermentations using GC-MS.


Screening | Cell Biology
I began my scientific career as a cancer biologist and have since gained extensive experience in the discovery and characterization of natural products from various origins (plants, semi-synthetic sources, and marine fungi) with applications as anticancer or antimicrobial agents.
- Human cancer cell culture (adherent and suspension cell lines).
- Development of protocols and bioassays for large-scale screening.
- Optimization and miniaturization of protocols (96-well plates).
- Cytotoxicity assays (manual methods and commercial kits) for anticancer activity discovery and identification of cell death mechanisms.
- Screening of fungal fermentations for enzymatic activity (e.g., lignocellulolytic, laccase, and peroxidase activities).
- Quantitative real-time PCR (qRT-PCR).
- Flow cytometry.
- Fluorescence microscopy.
- SDS-PAGE and Western blotting.
Synthetic Biology
After mining the genomes of several specimens from my Basidiomycota fungal strain library to isolate genes of interest, I have developed strong expertise in the isolation, cloning, and expression of genes with industrial and economic significance in appropriate heterologous hosts.
- DNA/RNA isolation from fungal cultures.
- Polymerase chain reaction (Touchdown PCR, Gradient PCR, Colony PCR, Multiplex PCR, 2-step PCR).
- Isolation of genes of interest from unsequenced and unique fungi.
- Sequence design, DNA assembly, and plasmid creation.
- Molecular cloning (restriction enzyme-free) of large and/or multiple fragments using Gibson assembly.
- Development, optimization, and simplification of complex protocols and SOPs.
- Yeast homologous recombination.
- Constitutive or inducible expression of intracellular or secreted proteins and biocatalysts.
- Expression in appropriate model and non-model organisms, including Escherichia coli, Pseudomonas putida, Deinococcus radiodurans, Bacillus subtilis, and the yeast Pichia pastoris.
- Strain engineering and metabolic pathway engineering.


Bioinformatics
The various research projects in molecular biology and biochemistry I have worked on have led me to develop strong bioinformatics and computational biology skills, covering different aspects, such as:
- BLAST and sequence alignment.
- Phylogenetic analysis.
- Genome mining, identification, and description of biosynthetic gene clusters.
- Gene prediction for Basidiomycota phylum.
- Python 3 programming and the BioPython library.
- Protein structure modeling and prediction (PyMOL, SWISS-MODEL, AlphaFold).
- Experience with various software (Aliview, CLC Genomics Workbench, Jalview, MEGA, SnapGene, UGENE).
- Experience with Linux (Ubuntu).
Coding | Programming
I recently decided to learn and develop coding and programming skills in R, Python 3, HTML5, and CSS3. Using online resources, my objective was to develop new skills that would help automate recurring tasks, solve complex professional challenges, manage and visualize large datasets, and generate high-quality figures.
R Programming Language (R Studio)
- Data management and analysis of large datasets.
- Data visualization (plots, gene clusters, etc.) and design of high-quality figures for publications and presentations.
Python 3 Programming Language
- Creation and development of smart Python 3 scripts and programs to solve challenging and complex problems, automate time-consuming workflows, and manage and analyze large datasets for biotechnology research projects (e.g., extraction of large datasets from files or online resources and websites).
Python 3 scripts and programs I have completed and am currently working on have been deposited on my personal GitHub profile page.
HTML5 - CSS3 - Bootstrap 5
- Design of this responsive website from scratch using HTML5 and CSS3.

Basidiomycota strains isolated
Cloning reactions
Mentees
(UG - PostDoc)
Fungi-themed clothing
Soft skills
Team Worker
Problem Solver
Self-Learner
Effective Communicator
Work Ethic
Innovator
Organized
Attention to Detail
My Research
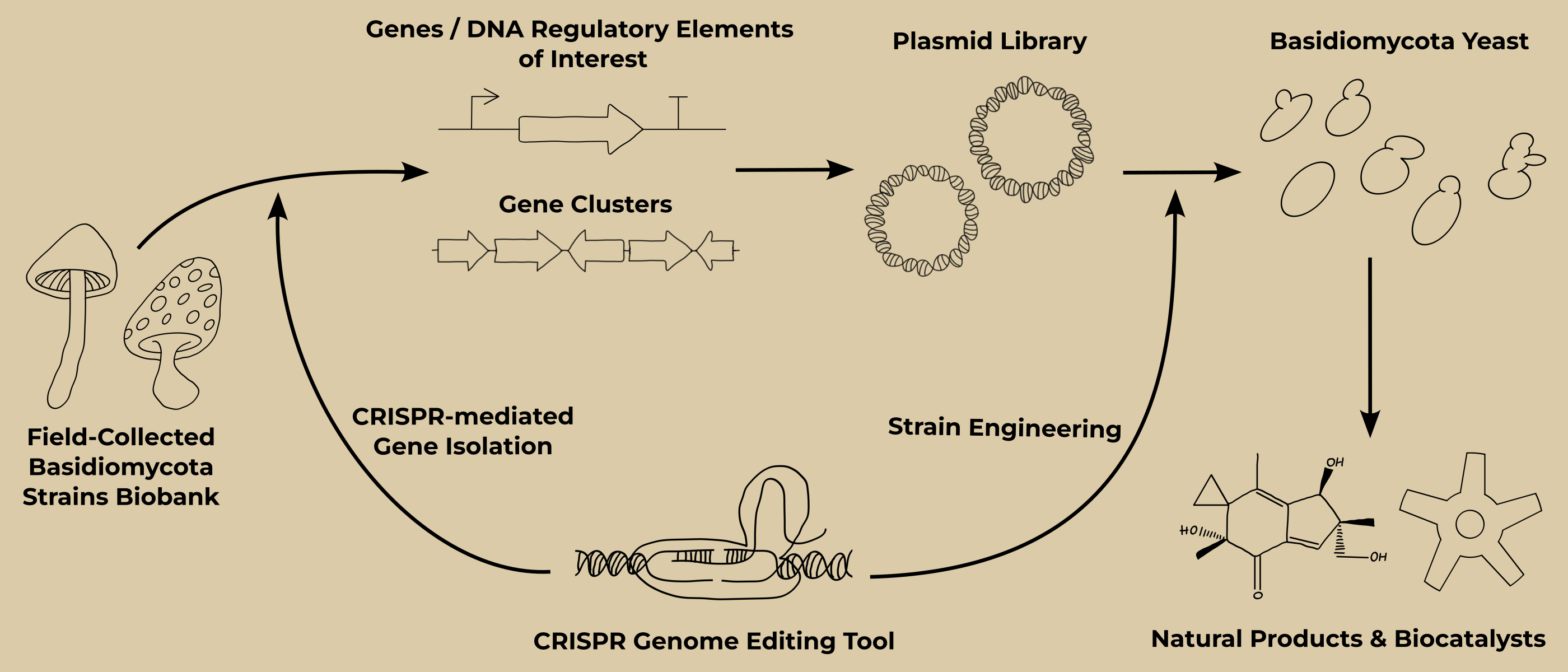
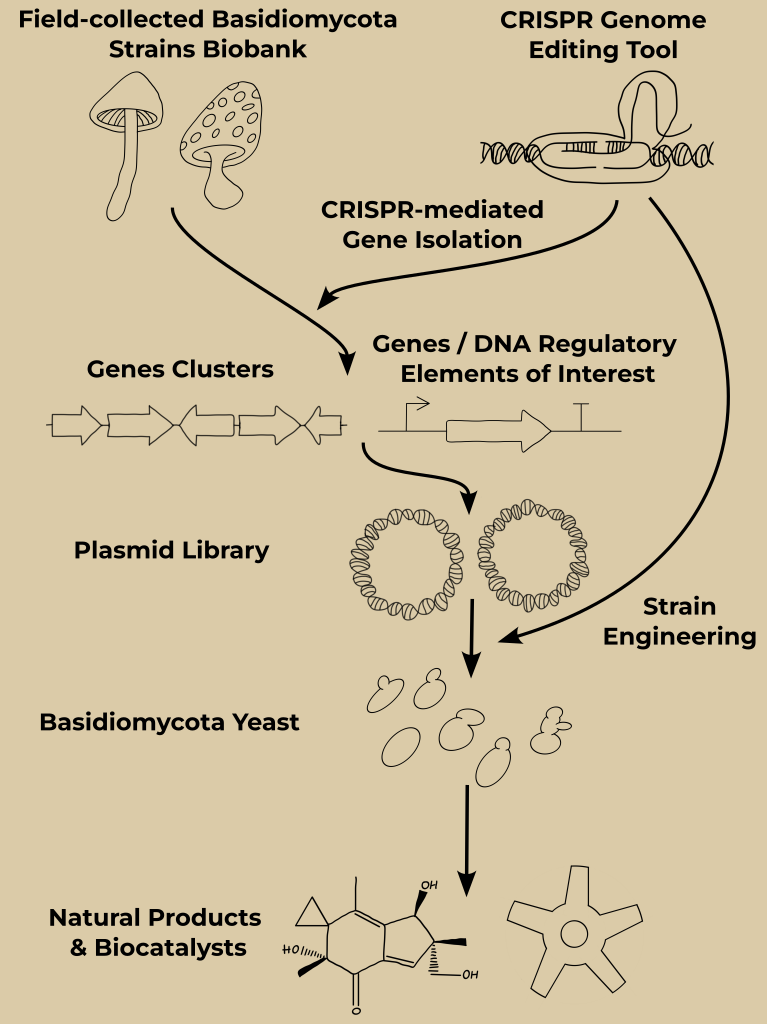
Development and engineering of a Basidiomycota yeast as a chassis for the heterologous expression of genes and gene clusters isolated from an unsequenced field-to-lab Basidiomycota fungi strain library.
Fungi, including Basidiomycota, have been used for centuries by humans as food and for their therapeutic properties in traditional medicines (1). Recent discoveries have highlighted and confirmed the presence of unique and novel bioactive natural products that are currently under investigation for the development of next-generation therapeutics (anticancer drugs, antibiotics, etc.) and unique biocatalysts with specific and varied activities that can provide eco-friendly solutions to current and future challenges (green chemistry, bioremediation, space exploration,...) (2, 3).
One reason for the biosynthesis of diverse and unique biomolecules by fungi might be explained by the long genetic diversity that began over 1 billion years ago (4). Moreover, fungal genetics is highly dynamic and plays a significant role in the biodiversity of the fungal kingdom, even at the intra-species level. Recent observations have shown that fungal specimens from the same species but different strains, collected in the same areas, exhibit a differential metabolome, resulting in the production of various natural products and biocatalysts, making each fungal strain unique in terms of secondary metabolites and biocatalysts produced (5).
However, the flip side of dynamic fungal genetics is what makes fungi difficult to work with. In addition to being challenging or impossible to grow under laboratory conditions, fungi have slow growth, resulting in limited biological material and biomolecules of interest. Moreover, fungi have evolved differently from other organisms, developing their own complex genetic organization and regulation (e.g., RNA splicing, epigenetics), as well as specific post-translational modifications (PTMs) (6), which further complicates their study. As a result, heterologous expression in non-fungal models is often used to bypass these limitations. This also highlights the need to identify and develop new tools for non-fungal models to enable efficient biosynthesis of bioactive compounds and biocatalysts.
Despite the availability of several heterologous host systems (insects, bacteria, ascomycota, yeast), the number of Basidiomycota chassis engineered for the biosynthesis of bioactive natural products and biocatalysts is still very limited (Agaricus bisporius, Coprinus cinereus, Armillaria mellea) and these systems have multiple limitations (e.g., different codon usage, slow growth, time-consuming protocols) (7).
Therefore, I am currently working on the development of a Basidiomycota yeast chassis for the expression of genes of interest and biosynthetic gene clusters:
- Development of an easy-to-transform robust Basidiomycota yeast chassis:
- Design and construction of a plasmid library for inducible or constitutive expression of genes of interest.
- Development and optimization of an efficient, easy, fast and low-cost transformation protocol.
- Engineering of the Basidiomycota yeast strain for easy-cloning :
- Cloning of markers and selection by Cre-Lox system.
- Specific and targeted genome editing by CRISPR technology.
- Knock-out/Knock-in of genes by CRISPR/Cas9 technology to alleviate bottlenecks and improve biosynthetis and expression of molecules of interest.
- Evaluation of the engineered Basidiomycota yeast strain for heterologous expression:
- Cloning and expression of single genes of interest (laccases, terpene synthases, sesquiterpene synthases, unspecific peroxygenases...).
- Cloning and expression of selected biosynthetic gene clusters from Basidiomycota using CRISPR/Cas9 system.
- Activation of silenced biosynthetic gene clusters by CRISPRa technology.
Publications
Molecular Identification and Antimicrobial Activity of Foliar Endophytic Fungi on the Brazilian Pepper Tree (Schinus terebinthifolius) Reveal New Species of Diaporthe
Germana D Dos Santos, Renata R Gomes, Rosana Gonçalves, Gheniffer Fornari, Beatriz HLNS Maia, Claudia Schmidt-Dannert, François Gaascht, Chirlei Glienke, Gabriela X Schneider, Israella R Colombo, Juliana Degenhardt-Goldbach, João LM Pietsch, Magda CV Costa-Ribeiro, Vania A Vicente
Current Microbiology 78.8 (2021): 3218-3229.
Discovery of Antifungal and Biofilm Preventative Compounds from Mycelial Cultures of a Unique North American Hericium sp. Fungus
Xun Song, François Gaascht, Claudia Schmidt-Dannert, Christine E. Salomon
Molecules 25.4 (2020): 963.
Molecular Identification and Antimicrobial Activity of Foliar Endophytic Fungi on the Brazilian Pepper Tree (Schinus terebinthifolius) Reveal New Species of Diaporthe
Weaam Ebrahim, Amal Hassan Aly, Victor Wray, Attila Mándi, Marie Hélène Teiten, François Gaascht, Barbora Orlikova, Matthias U Kassack, Wenhan Lin, Marc Diederich, Tibor Kurtan, Abdessamad Debbab, Peter Proksch
Journal of medicinal chemistry 56.7 (2013): 2991-2999.
DOI: 10.1021/jm400034b
Gallery










News & In the Media
News & Events

Minnesota State Fair 2025
Back at work at the Minnesota State Fair this year! Together with my colleagues, I had the chance to survey visitors about their opinions on how biotechnology can help tackle environmental challenges. This is part of our NSF (National Science Foundation)-funded project, which focuses on breaking down PFAS (“forever chemicals”) using engineered microorganisms.
Date: August 29, 2025
Location: Minnesota State Fair - University of Minnesota Driven to Discover Building (Falcon Heights, MN, USA)

University of Minnesota Day at the Capitol
As researchers from the University of Minnesota, my colleagues and I visited the Minnesota State Capitol to meet with local policymakers and advocate for Minnesota’s potential as a national hub for bioeconomy. We demonstrated how microorganisms, local Fungi and engineered Yeast, can help drive this transition. Fun fact: I was the first person to bring a bioreactor with 4L of Pichia pastoris to the Capitol!
Date: February 18, 2025
Location: Minnesota State Capitol (Saint Paul, MN, USA)

Minnesota State Fair 2024
I had the incredible opportunity to attend the Minnesota State Fair with my colleagues to present our research on biofilm engineering for bioremediation and biomining of rare earth elements (projects funded by the National Science Foundation) and to survey visitors about their perceptions of novel biotechnologies.
Date: August 29, 2024
Location: Minnesota State Fair - University of Minnesota Driven to Discover Building (Falcon Heights, MN, USA)

1-Day Symposium - Fungi: A Global Nexus for Cross-Kingdom Interactions
I attended the symposium "Fungi: A Global Nexus for Cross-Kingdom Interactions" (Website) organized by the Department of Plant Pathology (University of Minnesota) and I had the opportunity to present my recent work on my Basidiomycota Fungi research project.
Date: September 15, 2023
Location: University of Minnesota - Cargill Building (Saint Paul, MN, USA)

Society for Industrial Microbiology and Biotechnology - Annual Conference
I was awarded a BTI (BioTechnology Institute, University of Minnesota, USA) Travel Grant and I attended the SIMB (Society for Industrial Microbiology and Biotechnology) Annual Conference (Website) in Minneapolis (Minnesota, USA) and I had the opportunity to present my recent work on strain engineering of the radiation resistant microorganism Deinococcus radiodurans.
Date: July 30 - August 2, 2023
Location: Hyatt Regency Hotel (Minneapolis, MN, USA)
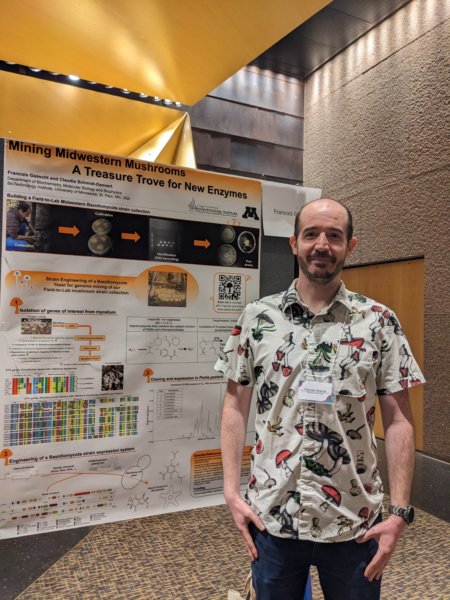
The 1st North America-Japan Enzyme Technology Symposium
I attended the 1st North America-Japan Enzyme Technology Symposium (Website) organized by Amano and the BioTechnology Institute (University of Minnesota, MN, USA) and I presented a poster about my work on the Basidiomycota project.
Date: May 5, 2023
Location: The University of Minnesota - McNamara Alumni Center (Minneapolis, MN USA.)

Lab visit by the CORE Scholars
Visit of the Schmidt-Dannert Laboratory by a group of students and parents of the Community Outreach Retention and Engagement (CORE) Program. I had the opportunity to show some microbial cultures, including some Fungi I collected to the CORE Scholars and their families.
To know more about the CORE Program Website
Date: March 18, 2023
Location: Schmidt-Dannert Laboratory (Saint-Paul, MN USA.)
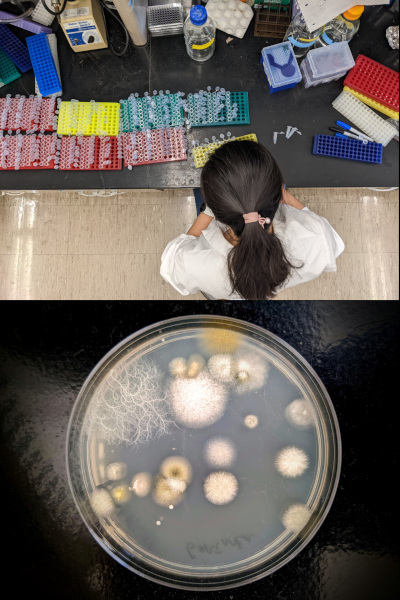
BioTechnology Institute - Summer Photo Contest
Two of my pictures were awarded during the Summer Biotechnology Institute Photo Contest.
Top: Colleague working on biological samples.
Bottom: Petri dish with LB medium was exposed during 1 minute under a tree in front of Gortner Laboratory (Minnesota, USA) and incubated for 72 hours at 30°C.
Date: November 10, 2022
Location: BioTechnology Institute - Gortner Laboratory of Biochemistry (Saint-Paul, MN USA.)
Press

Principles and Applications of Soil Microbiology
My culture of mycelium (Coprinopsis strossmayeri) on malt extract agar was used in Chapter 6 - Fungi (Figure 6.3) to illustrate the pattern of growth of Fungi mycelium.
2021
read
Il y a tellement de choses à visiter au Luxembourg
Article published by Paperjam.lu, a Luxembourg-based media about my expatriation and my life vision from Minnesota.
August 2019
read
The best ambassador for Luxembourg… can be anyone
Article published by Delano Magazine, an independant Luxembourg-based media when I was elected as one of the 100 personalities who illuminate Luxembourg's brand image abroad and contribute positively to Luxembourg's international reputation.
November 2017
read
BIO INSPIRED
One of my mushroom (Polyporus squamosus) culture was selected for cover of BIO INSPIRED, a publication for alumni, donors and friends of the College of Biosciences at the University of Minnesota (USA).
Spring 2017
read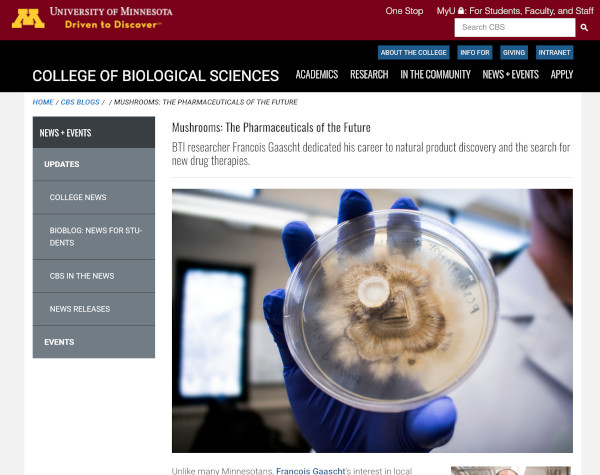
Mushrooms: The Pharmaceuticals of the Future
Article published by the College of Biological Science - University of Minnesota about my Fungi research project at the BioTechnology Institute.
April 2017
readContact Me
I am always happy to connect with people to exchange ideas, share my expertise, and assist with your next challenging project. Feel free to reach out via email or connect with me on social media.